Prioritization Component (BiERapp)¶
Access¶
BiERapp is available at the following address.
Note
The address is accessible only via VPN.
BiERapp¶
** This tool is aimed at discovering new disease genes or variants by studying affected families or cases and controls. It carries out a filtering process to sequentially remove: (i) variants which are not compatible with the disease because they are not expected to have impact on the protein function; (ii) variants that exist at frequencies incompatible with the disease; (iii) variants that do not segregate with the disease. The result is a reduced set of disease gene candidates that should be further validated experimentally. **
BiERapp (28) efficiently helps in the identification of causative variants in family and sporadic genetic diseases. The program reads lists of predicted variants (nucleotide substitutions and indels) in affected individuals or tumor samples and controls. In family studies, different modes of inheritance can easily be defined to filter out variants that do not segregate with the disease along the family. Moreover, BiERapp integrates additional information such as allelic frequencies in the general population and the most popular damaging scores to further narrow down the number of putative variants in successive filtering steps. BiERapp provides an interactive and user-friendly interface that implements the filtering strategy used in the context of a large-scale genomic project carried out by the Spanish Network for Research, in Rare Diseases (CIBERER), and the Medical Genome Project. in which more than 800 exomes have been analyzed.
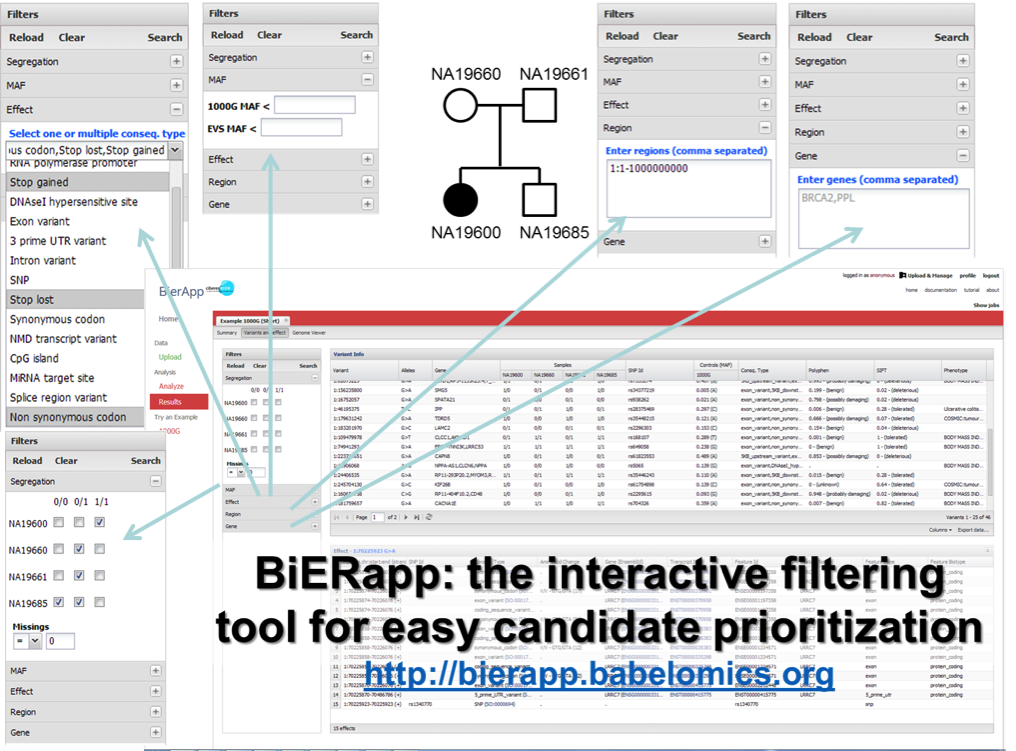
Figure 6. Web interface to the prioritization tool. This figure shows the interface of the web tool for candidate gene prioritization with the filters available. The tool includes a genomic viewer (Genome Maps 30) that enables the representation of the variants in the corresponding genomic coordinates.